topGO
The analysis of gene expression data is an important tool for
understanding
mechanisms of living systems. Microarray experiments provide large
amounts of
data, but it is difficult to understand biological processes or
molecular functions from such data alone.
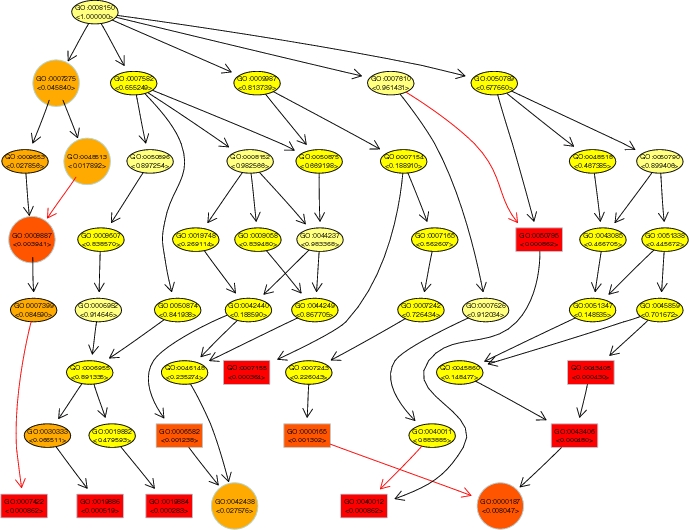
topGO (topology-based Gene
Ontology scoring) is a software package for calculating the
significance of biological terms from gene expression data. It
implements various standard
and advanced new algorithms for determining the relevance of Gene
Ontology groups from
microarrays.
A specific feature of the advanced algorithms is the exploitation of
the
hierarchical graph structure of the GO annotation for coping with the
large number
of GO groups. Often, related biological terms are scored with a similar
statistical significance.
Dependencies between GO terms can be de-correlated by accounting for
the neighborhood of
a GO node when calculating its significance. The new algorithms better
detect significant
GO terms from gene expression data.
topGO package provides tools for testing GO terms while accounting
for the topology of the GO graph. Different test statistics and
different methods for eliminating local similarities and dependencies
between GO terms can be implemented and applied.
Contact:
References:
- Wolfgang A Schulz, Adrian Alexa, Volker Jung, Christiane Hader,
Michele J Hoffmann, Masanori Yamanaka, Sandy Fritzsche, Agnes
Wlazlinski, Mirko Muller, Thomas Lengauer, Rainer Engers, Andrea R
Florl, Bernd Wullich, Jörg Rahnenführer
Factor
interaction analysis for chromosome 8 and DNA methylation alterations
highlights innate immune response suppression and cytoskeletal changes
in prostate cancer
Molecular Cancer, 6:14, 2007.
- Adrian Alexa, Jörg Rahnenführer, Thomas Lengauer
Improved
scoring of functional groups from gene expression data by decorrelating
GO graph structure
Bioinformatics, 13, 1600-1607, 2006.